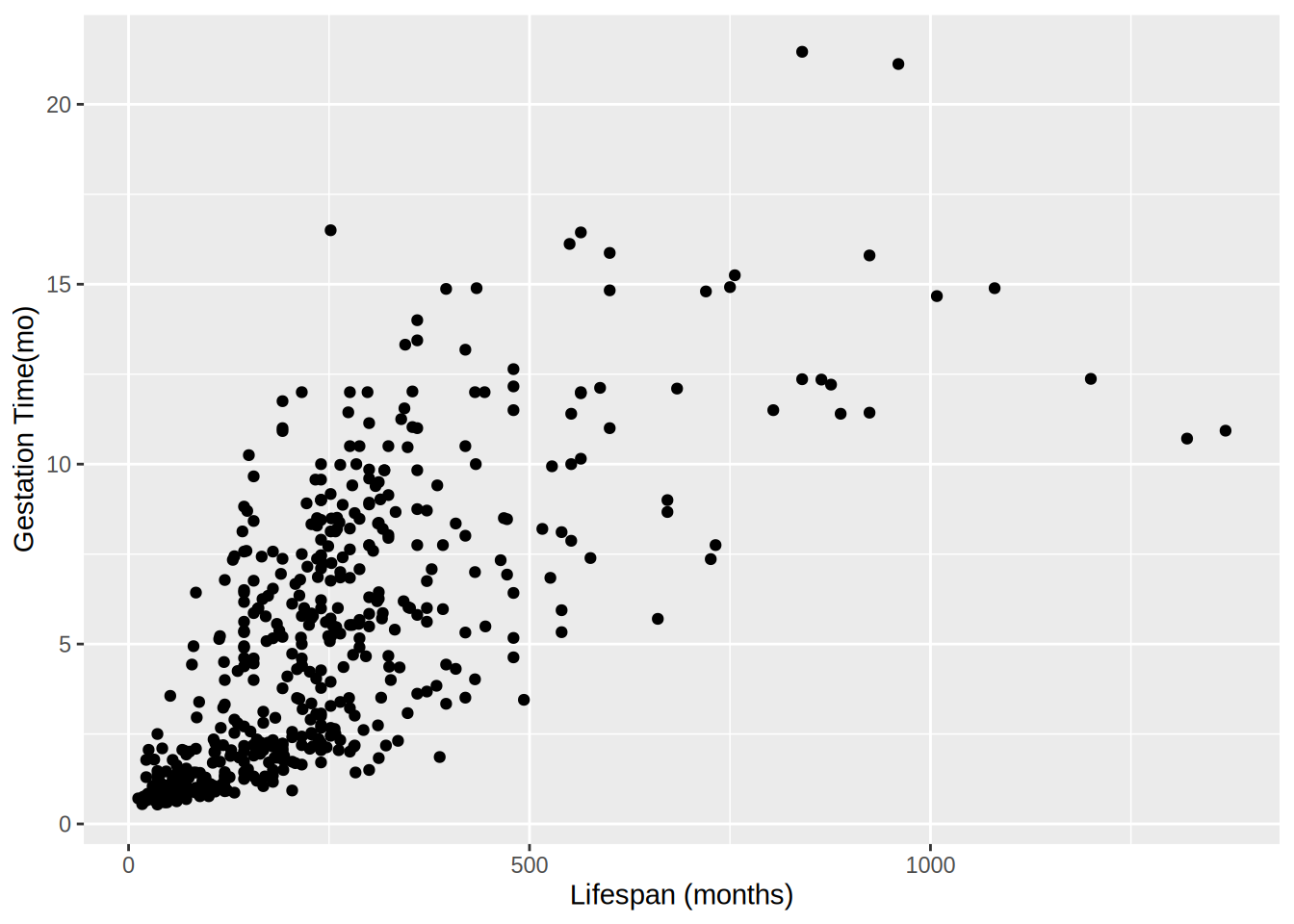
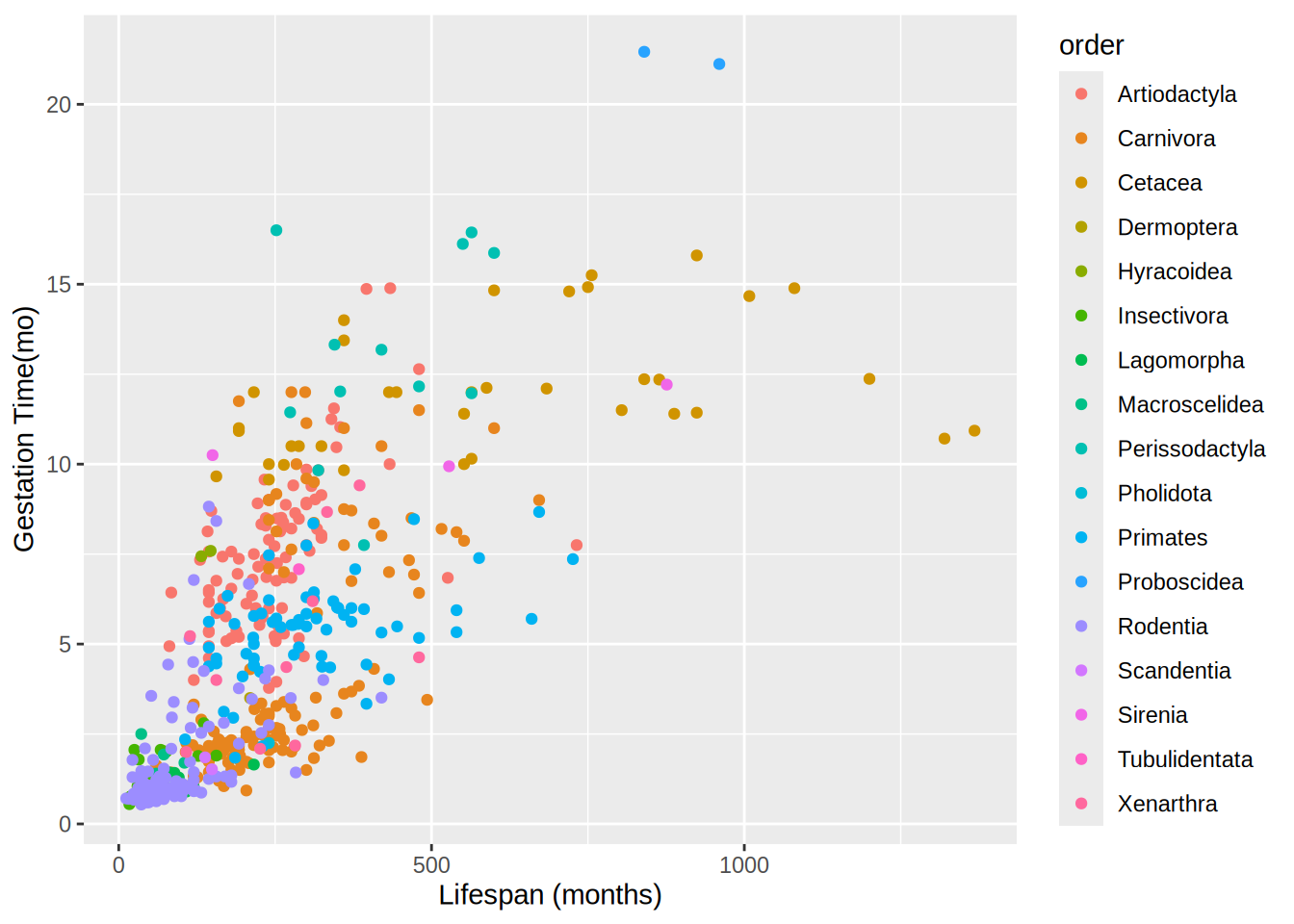
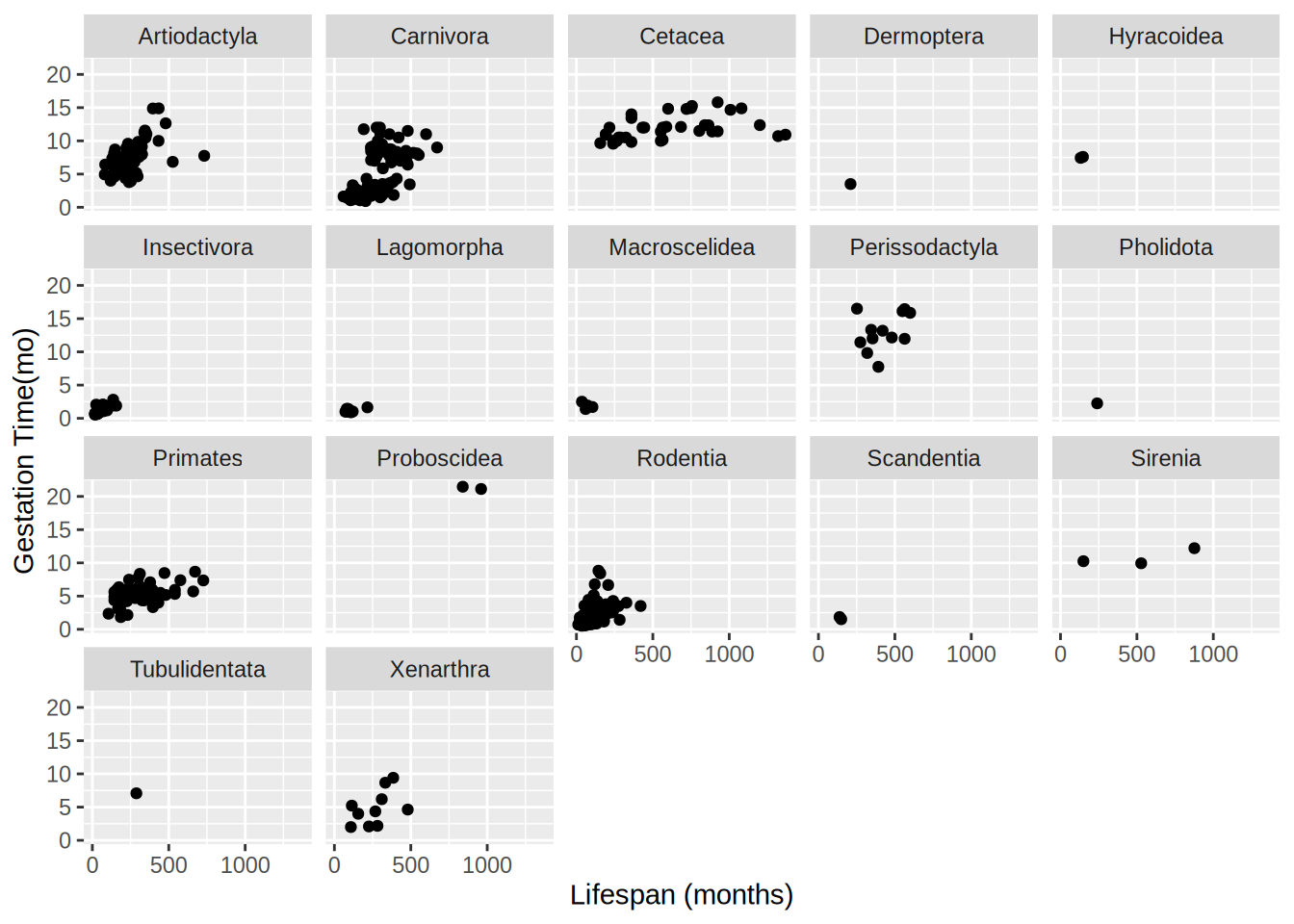
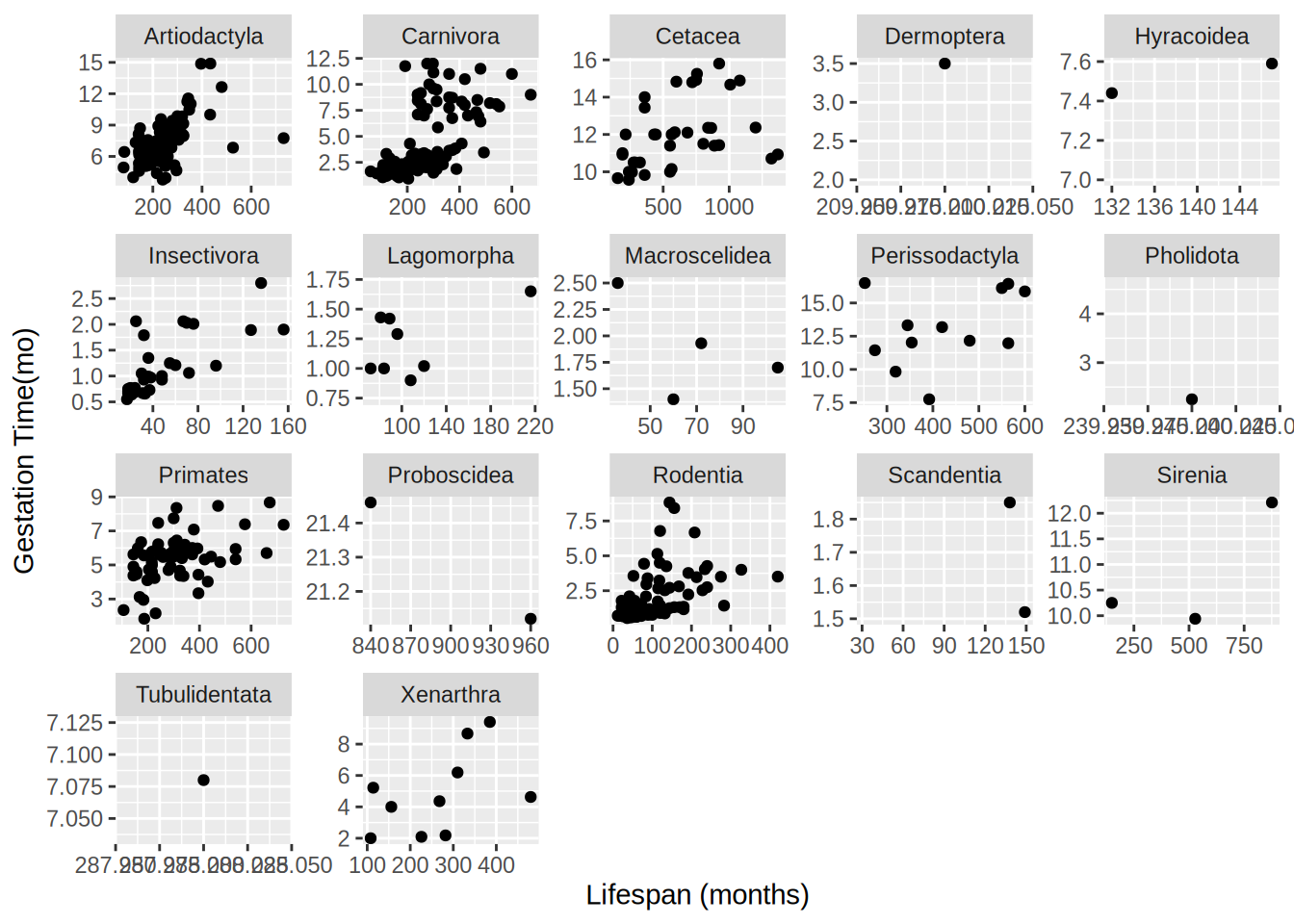
Attaching package: 'dplyr'The following objects are masked from 'package:stats':
filter, lagThe following objects are masked from 'package:base':
intersect, setdiff, setequal, unionRows: 1440 Columns: 14
── Column specification ────────────────────────────────────────────────────────
Delimiter: "\t"
chr (4): order, family, Genus, species
dbl (9): mass(g), gestation(mo), newborn(g), weaning(mo), wean mass(g), AFR(...
num (1): refs
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.