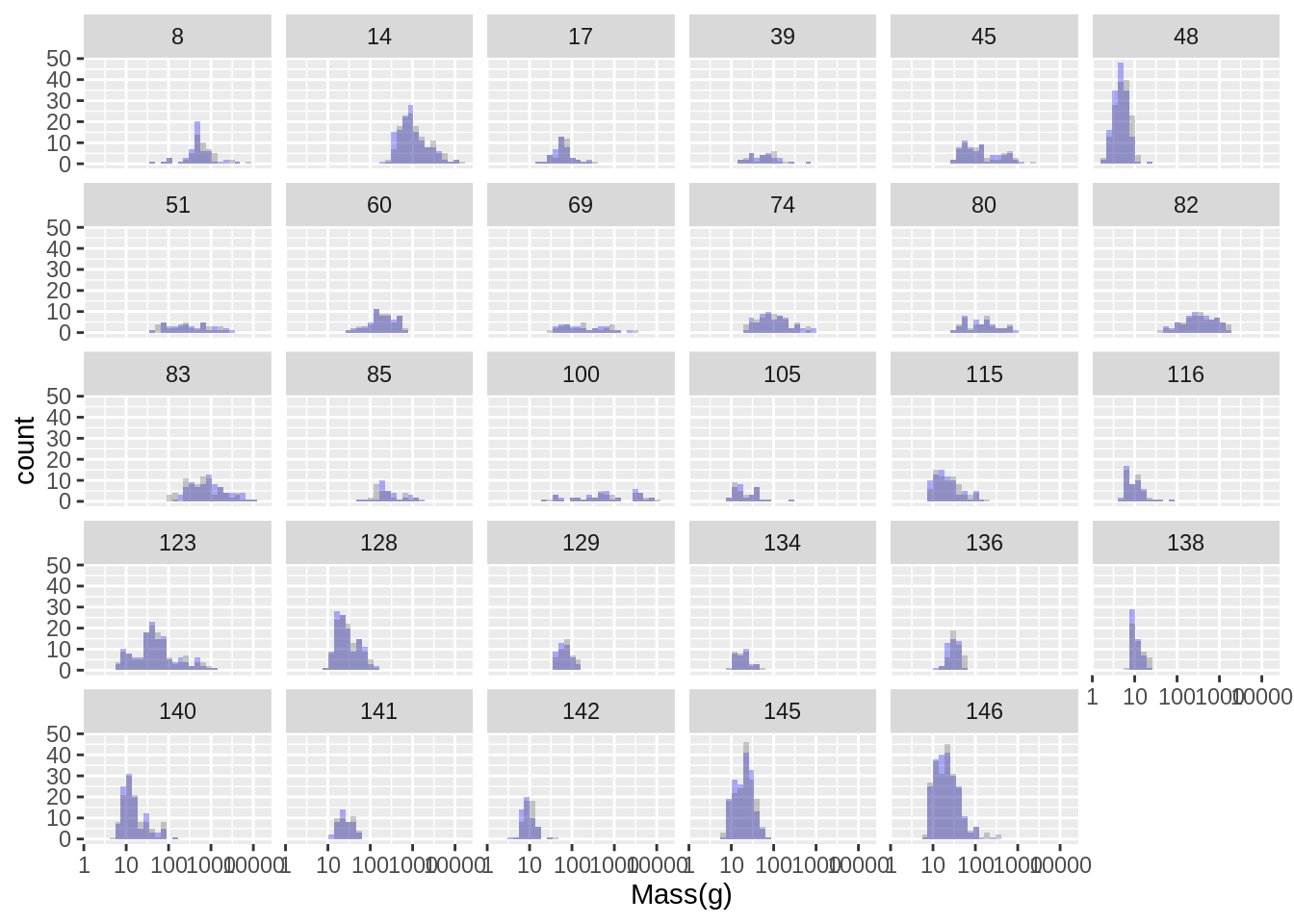
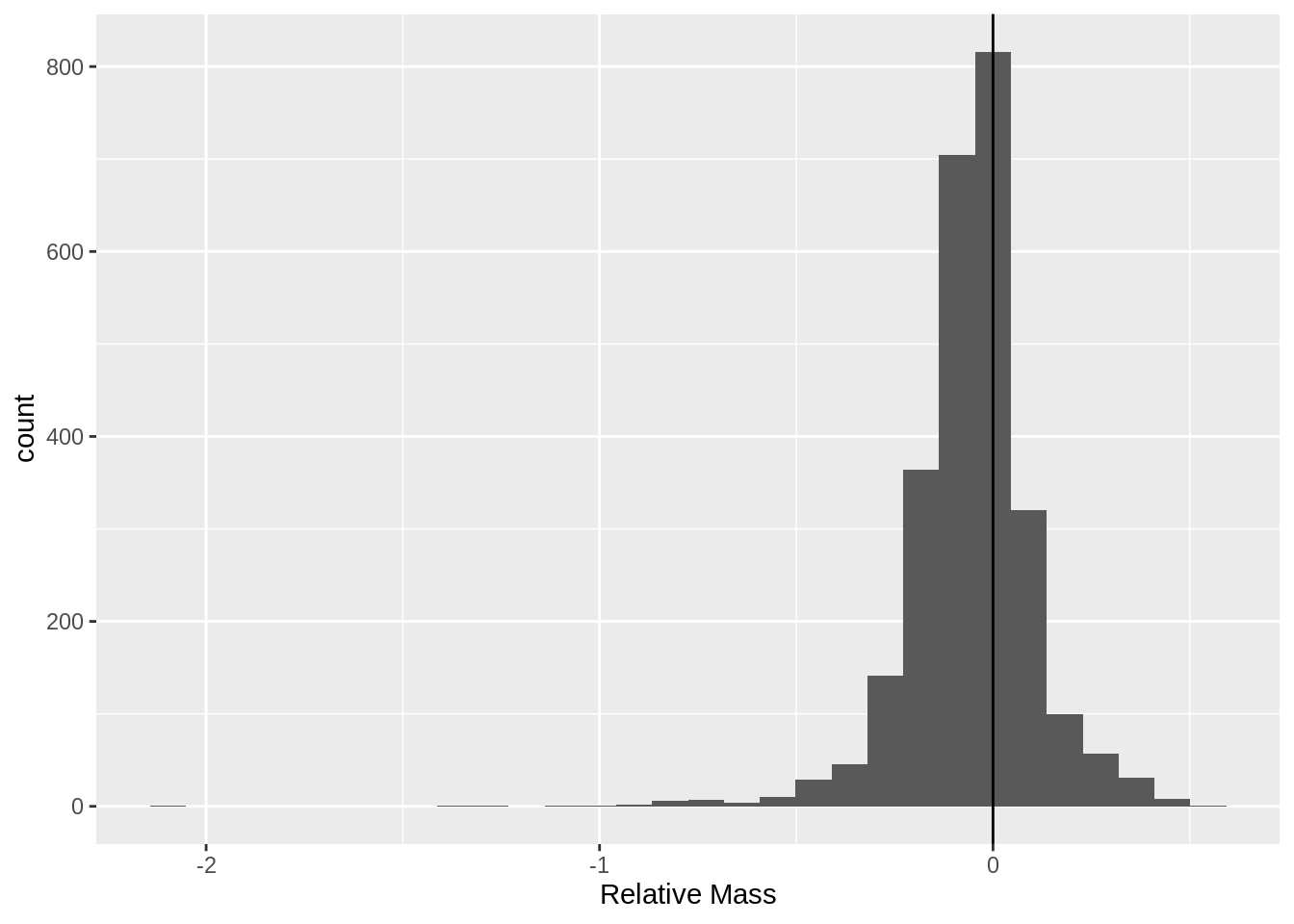
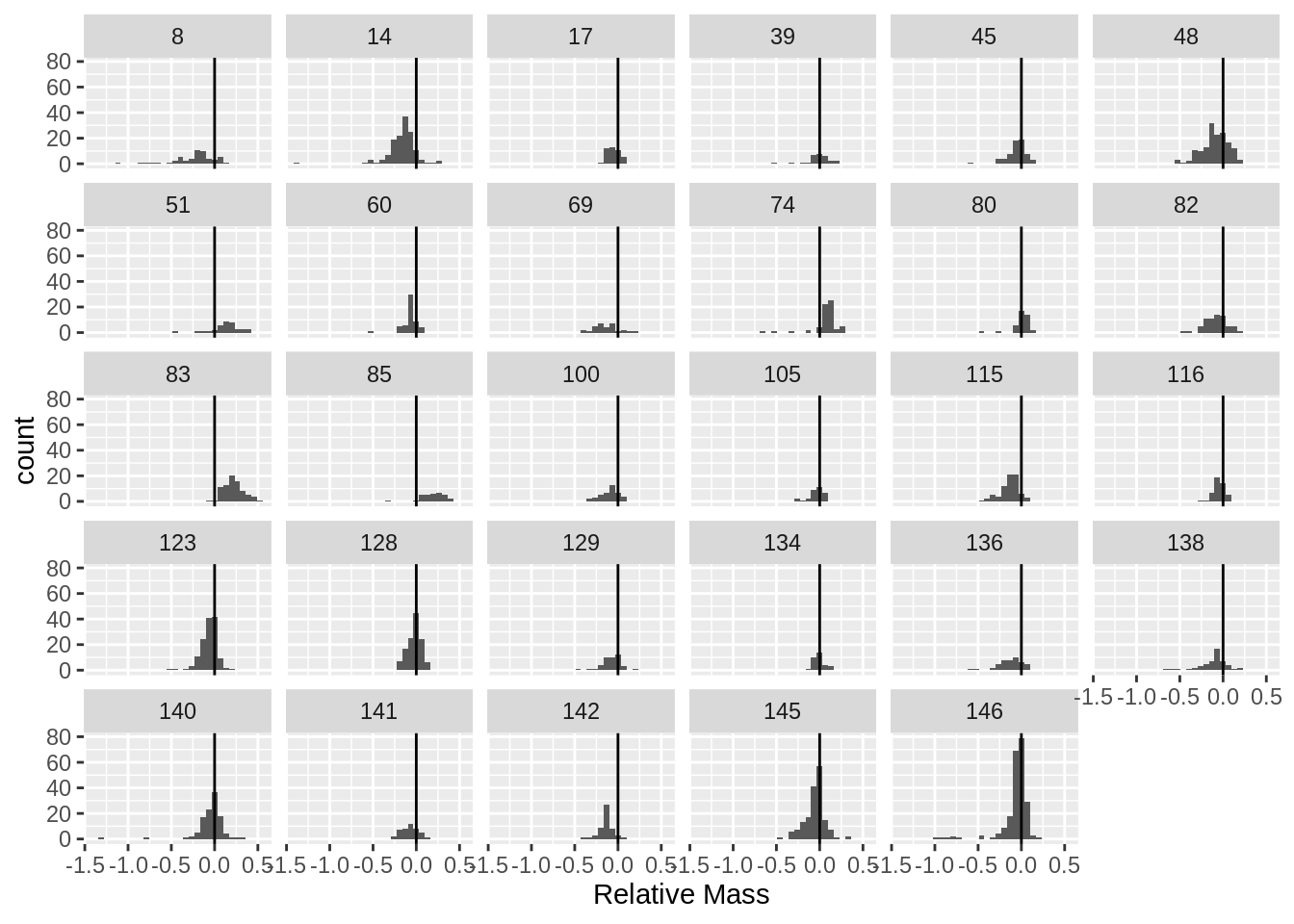
Attaching package: 'dplyr'
The following objects are masked from 'package:stats':
filter, lag
The following objects are masked from 'package:base':
intersect, setdiff, setequal, union
New names:
• `` -> `...42`
• `` -> `...43`
• `` -> `...44`
Warning: One or more parsing issues, call `problems()` on your data frame for details,
e.g.:
dat <- vroom(...)
problems(dat)
Rows: 3769 Columns: 44
── Column specification ────────────────────────────────────────────────────────
Delimiter: "\t"
chr (4): Species_name, English_name, Subspecies, References
dbl (37): Family, Species_number, M_mass, M_mass_N, F_mass, F_mass_N, unsexe...
lgl (3): ...42, ...43, ...44
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
`stat_bin()` using `bins = 30`. Pick better value `binwidth`.
`stat_bin()` using `bins = 30`. Pick better value `binwidth`.
`stat_bin()` using `bins = 30`. Pick better value `binwidth`.
Warning: Removed 1118 rows containing non-finite outside the scale range
(`stat_bin()`).
`stat_bin()` using `bins = 30`. Pick better value `binwidth`.