Attaching package: 'dplyr'
The following objects are masked from 'package:stats':
filter, lag
The following objects are masked from 'package:base':
intersect, setdiff, setequal, union
Rows: 5731 Columns: 8
── Column specification ────────────────────────────────────────────────────────
Delimiter: "\t"
chr (7): continent, status, order, family, genus, species, reference
dbl (1): mass
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
spc_tbl_ [5,731 × 8] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
$ continent: chr [1:5731] "AF" "AF" "AF" "AF" ...
$ status : chr [1:5731] "extant" "extant" "extant" "extant" ...
$ order : chr [1:5731] "Artiodactyla" "Artiodactyla" "Artiodactyla" "Artiodactyla" ...
$ family : chr [1:5731] "Bovidae" "Bovidae" "Bovidae" "Bovidae" ...
$ genus : chr [1:5731] "Addax" "Aepyceros" "Alcelaphus" "Ammodorcas" ...
$ species : chr [1:5731] "nasomaculatus" "melampus" "buselaphus" "clarkei" ...
$ mass : num [1:5731] 70000 52500 171002 28050 48000 ...
$ reference: chr [1:5731] "60" "63, 70" "63, 70" "60" ...
- attr(*, "spec")=
.. cols(
.. continent = col_character(),
.. status = col_character(),
.. order = col_character(),
.. family = col_character(),
.. genus = col_character(),
.. species = col_character(),
.. mass = col_double(),
.. reference = col_character()
.. )
- attr(*, "problems")=<externalptr>
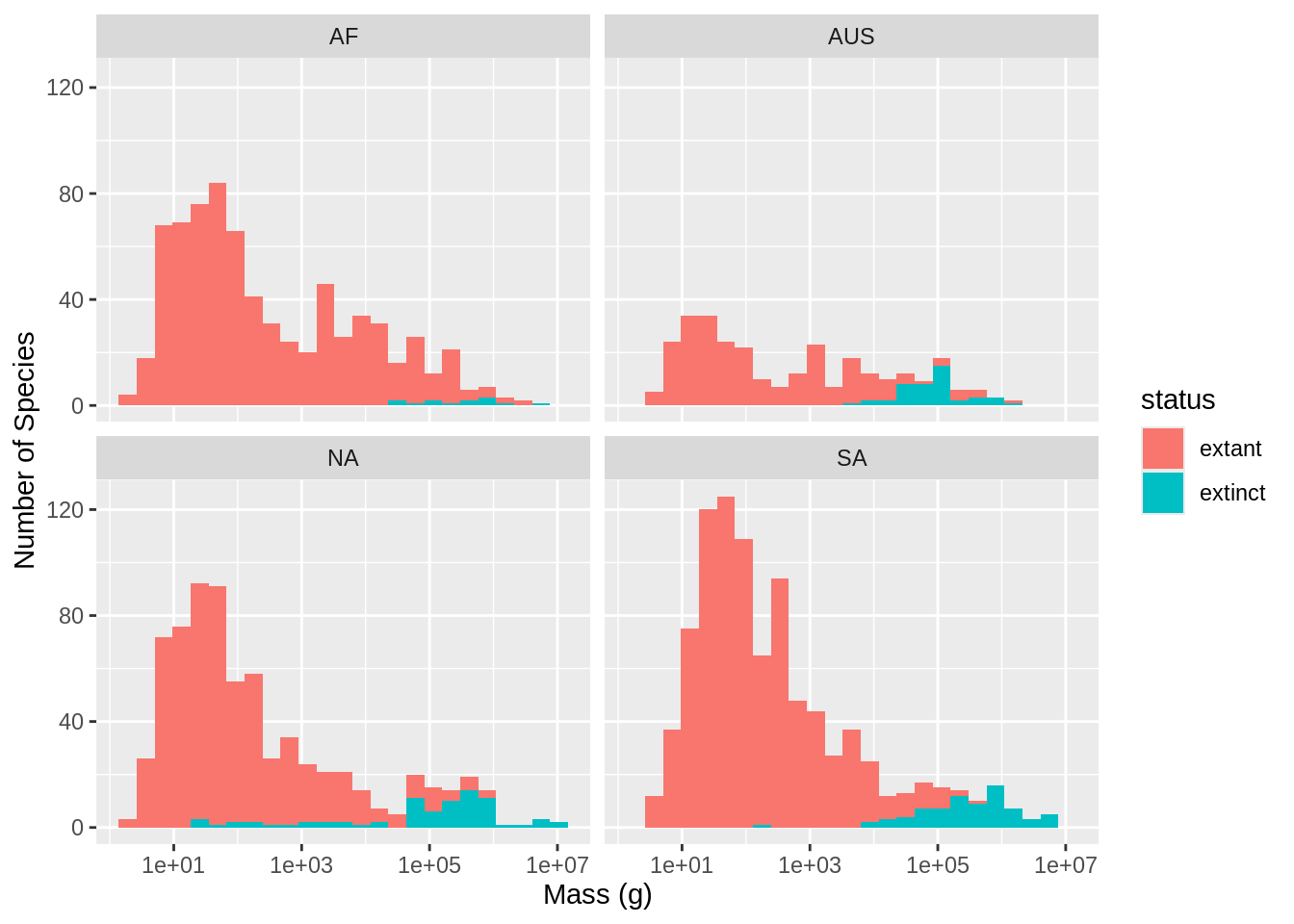
Figure 1
Warning: Removed 417 rows containing non-finite outside the scale range
(`stat_bin()`).
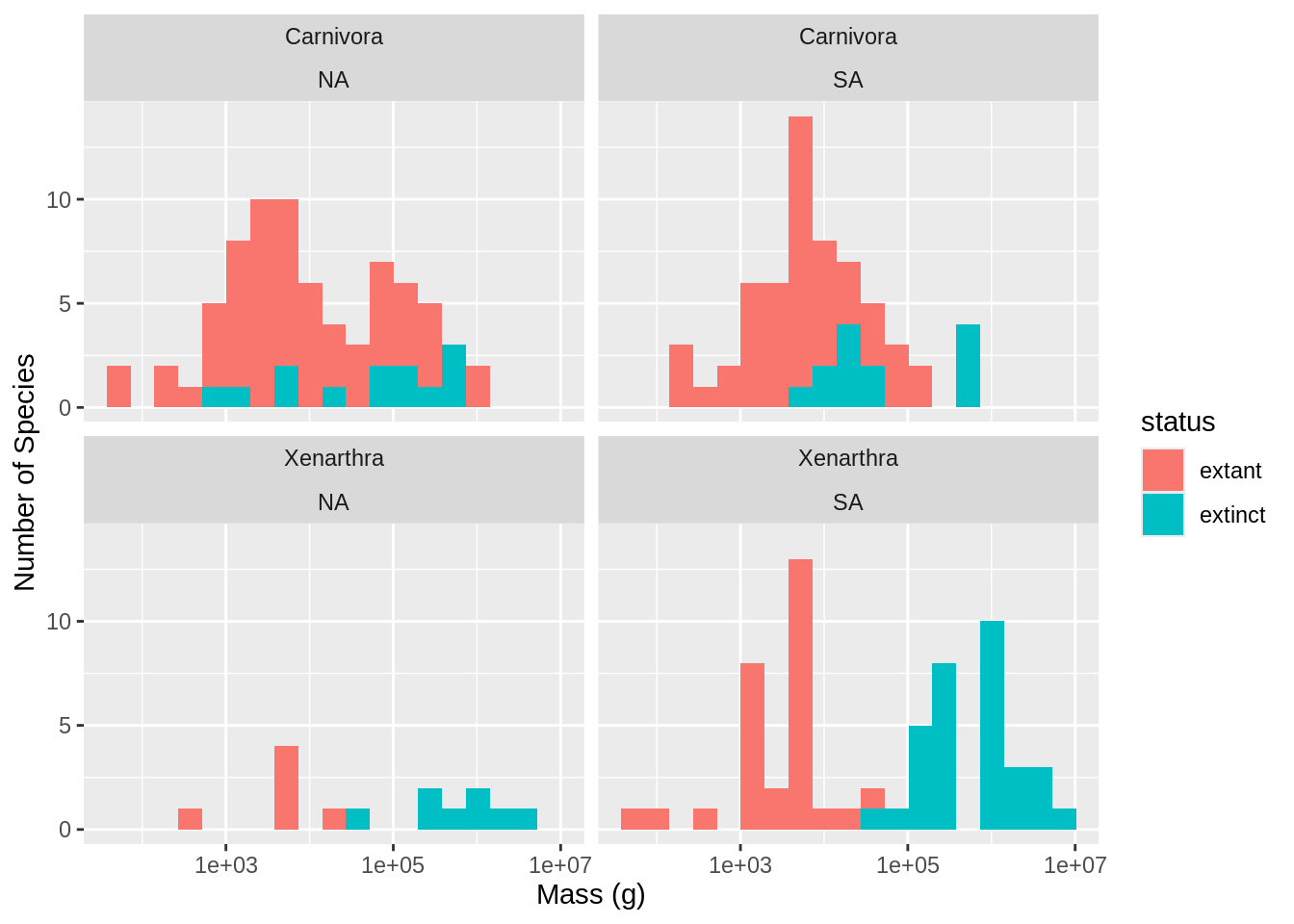
Figure 2
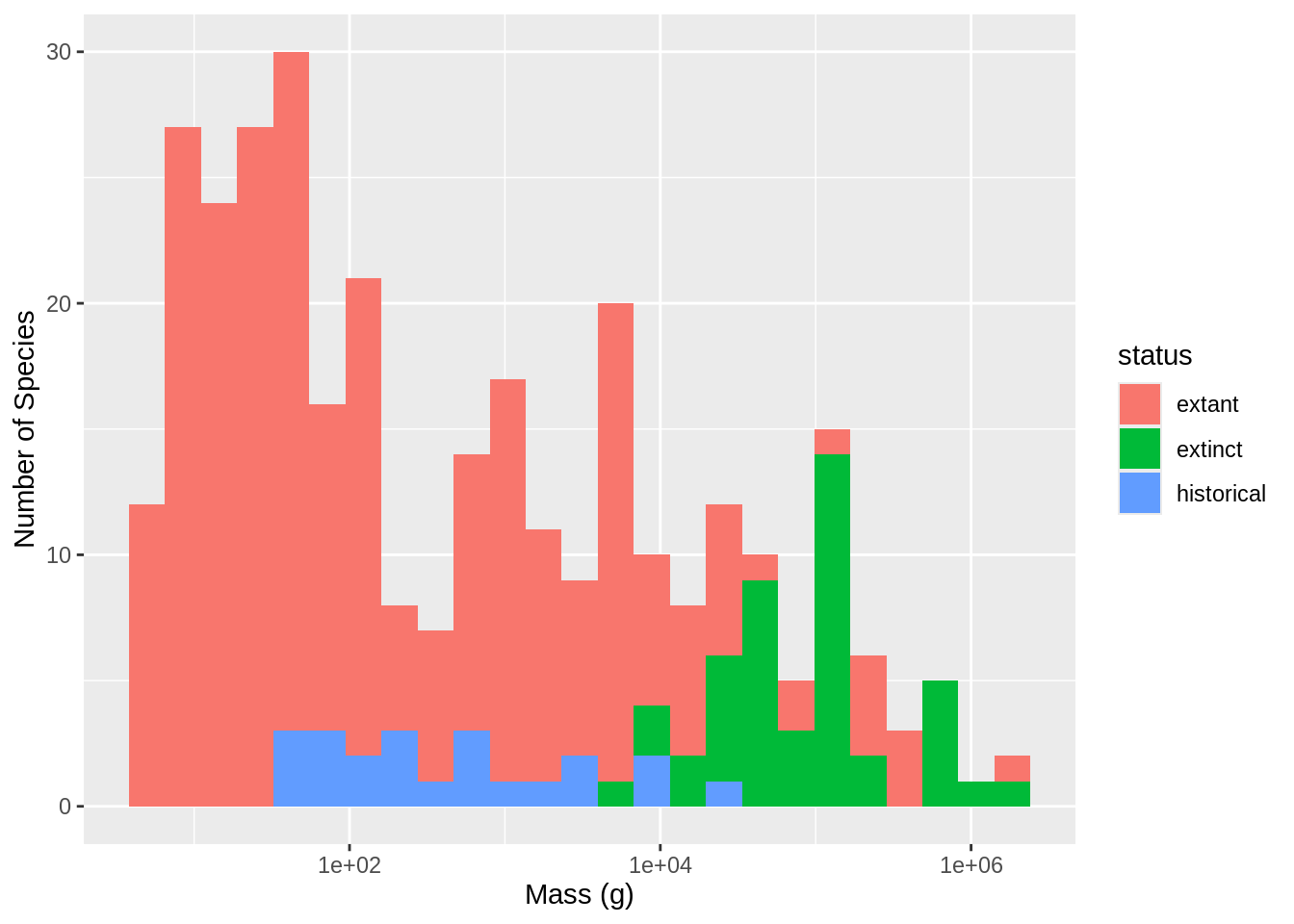
Figure 3
Warning: Removed 9 rows containing non-finite outside the scale range
(`stat_bin()`).
Optional This is a fancy way to dynamically eliminate sites with no extinct species But EA can also just be eliminated manually by adding it to the continent filtering step below.
Joining with `by = join_by(continent)`