Attaching package: 'dplyr'
The following objects are masked from 'package:stats':
filter, lag
The following objects are masked from 'package:base':
intersect, setdiff, setequal, union
Rows: 1440 Columns: 14
── Column specification ────────────────────────────────────────────────────────
Delimiter: "\t"
chr (4): order, family, Genus, species
dbl (9): mass(g), gestation(mo), newborn(g), weaning(mo), wean mass(g), AFR(...
num (1): refs
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
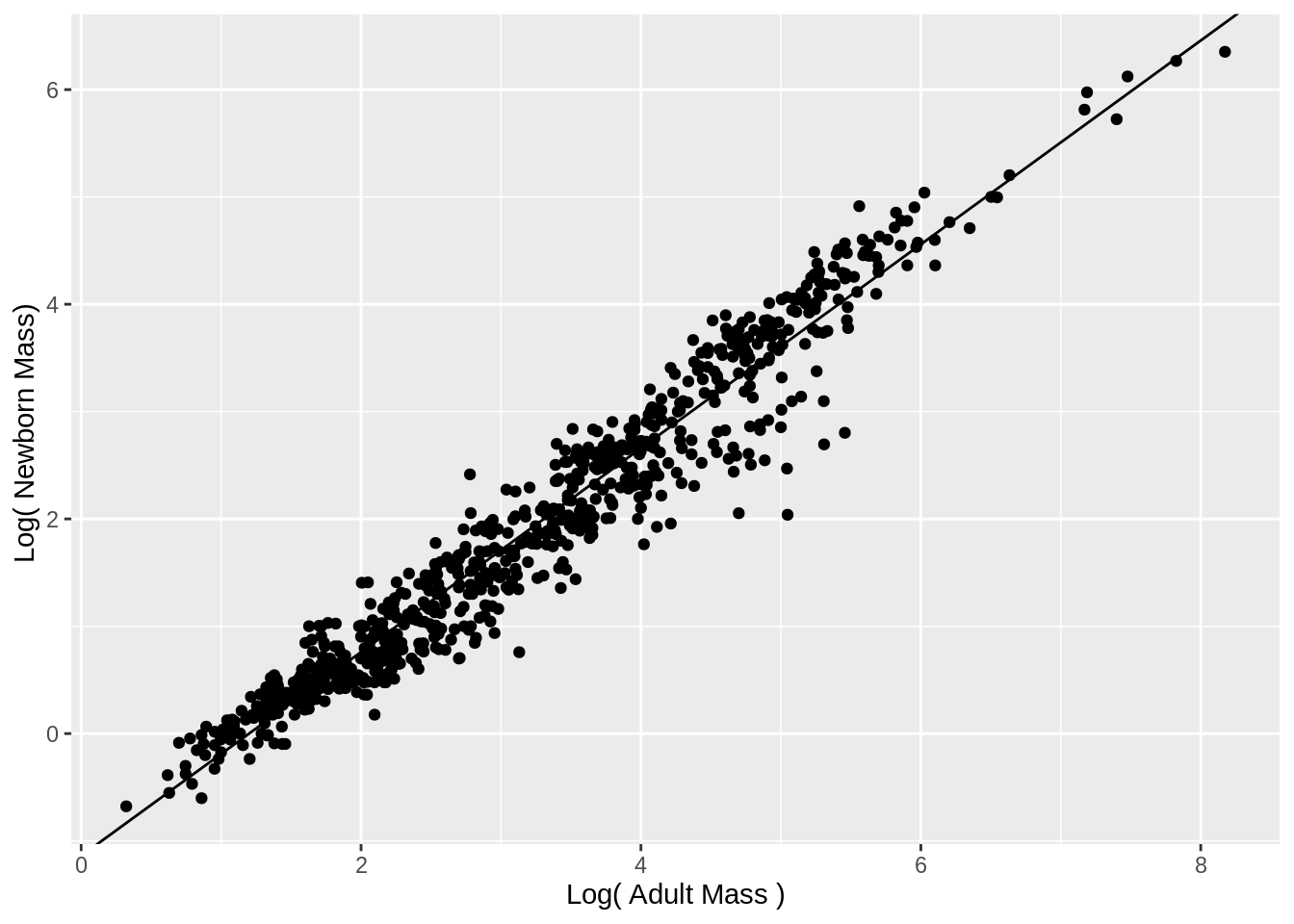
- Do a regression where x is log10(adult mass) and y is log10(newborn mass)
- Print the summary statistics for this regression
Call:
lm(formula = log10(`newborn(g)`) ~ log10(`mass(g)`), data = mammal_histories)
Residuals:
Min 1Q Median 3Q Max
-1.61535 -0.18339 0.04104 0.21687 0.91622
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -1.139431 0.027118 -42.02 <2e-16 ***
log10(`mass(g)`) 0.949620 0.007812 121.57 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.3155 on 814 degrees of freedom
(624 observations deleted due to missingness)
Multiple R-squared: 0.9478, Adjusted R-squared: 0.9477
F-statistic: 1.478e+04 on 1 and 814 DF, p-value: < 2.2e-16
- Make a graph.
Warning: `qplot()` was deprecated in ggplot2 3.4.0.
Warning: Removed 624 rows containing missing values or values outside the scale range
(`geom_point()`).
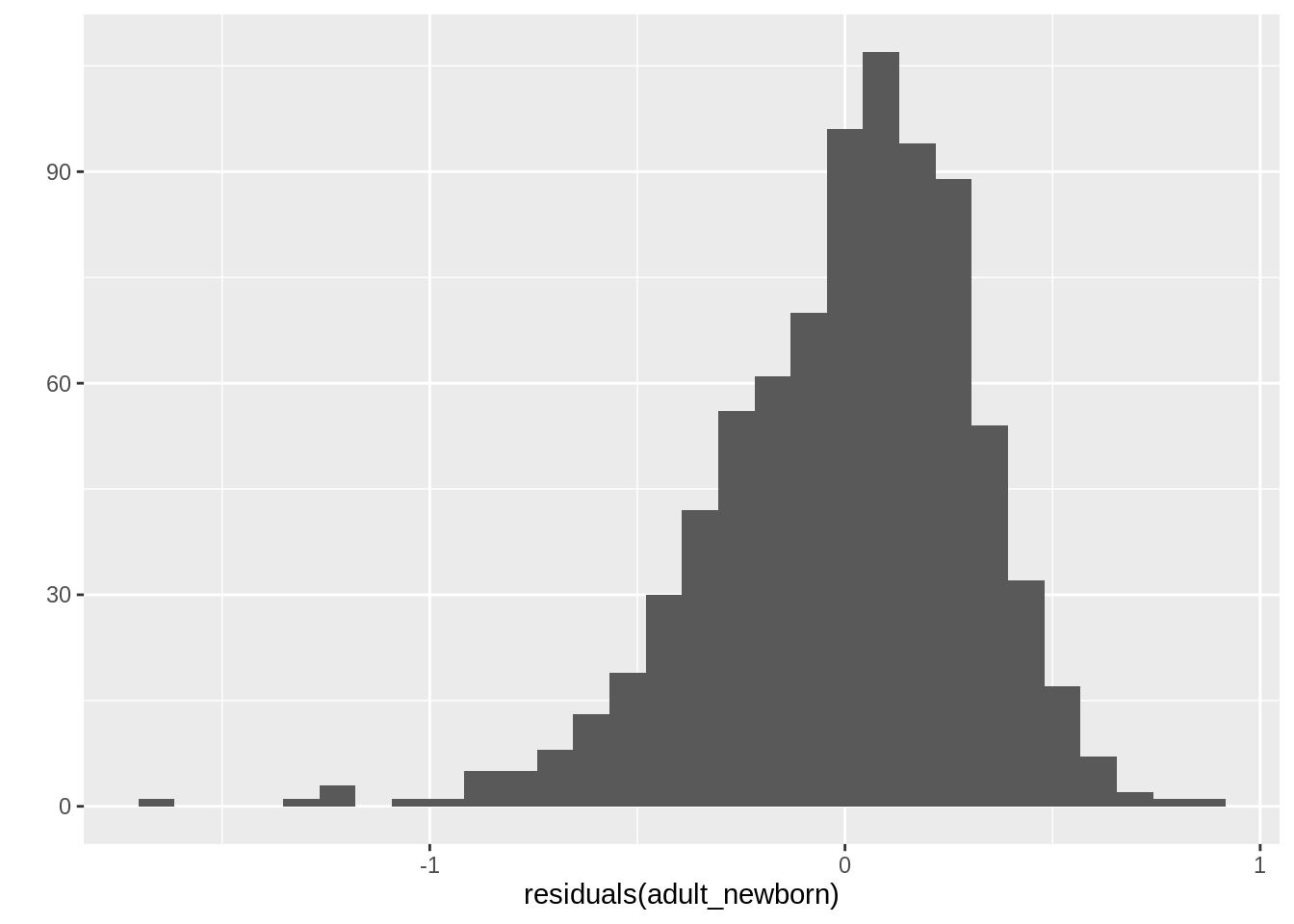
*Optional: plot a histogram of the residuals
`stat_bin()` using `bins = 30`. Pick better value `binwidth`.